Looks can deceive: Evolutionary trees based on anatomy may be wrong
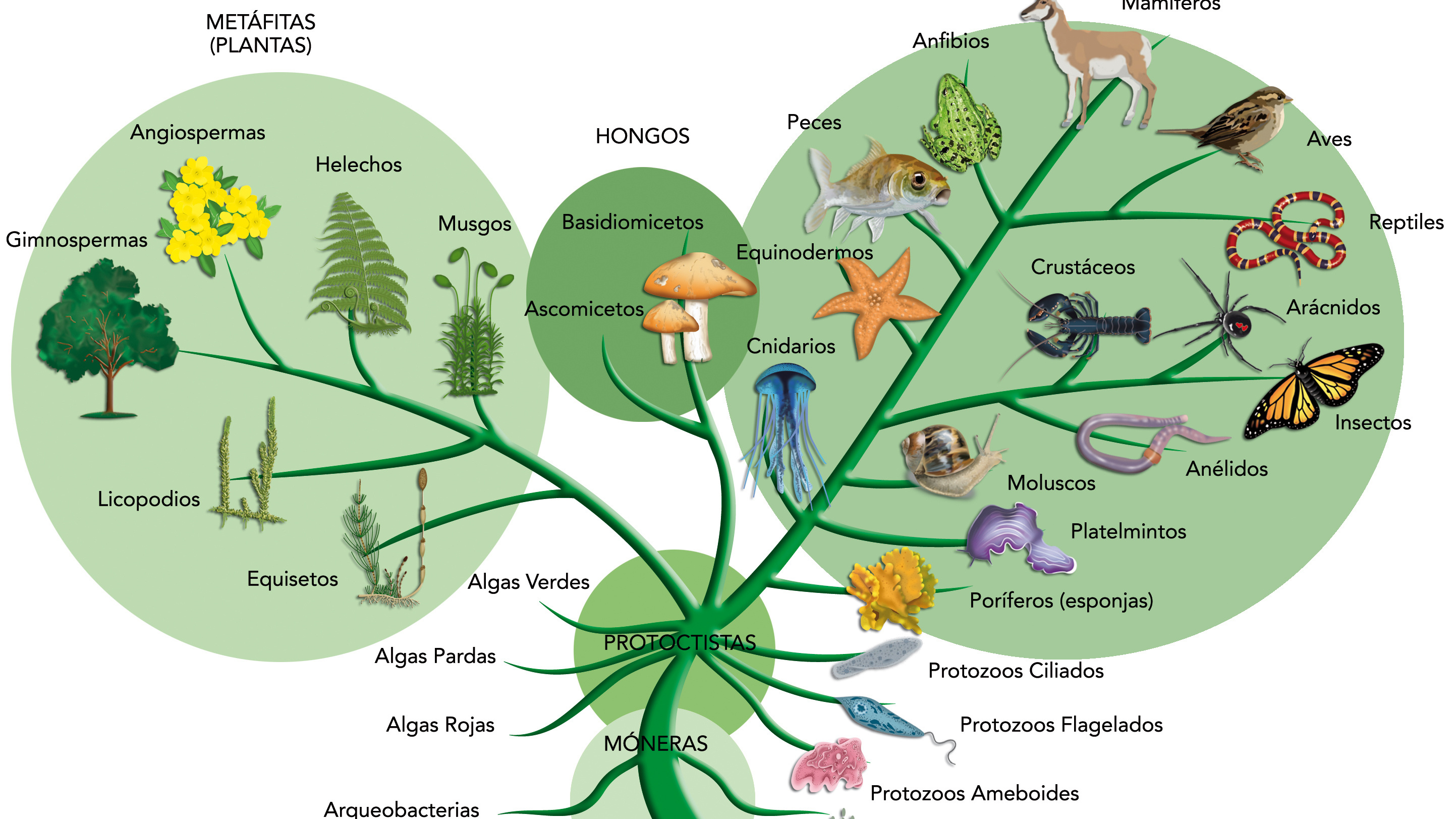
- Before the advent of DNA sequencing, biologists built evolutionary trees based on the similarity of organisms’ anatomy and structure. Now, molecular sequencing is used too, but there is no standard way of inferring relationships between organisms.
- A new study finds that phylogenetic trees based on DNA more closely follow species’ geographical distributions, a factor associated with evolutionary relationships.
- The data suggests that many trees built from morphology could be wrong, causing us to question many of our assumptions about relatedness and evolution.
The evolutionary relationships of Earth’s many living creatures have big implications for a range of fields within biology and ecology. It is therefore very important to map these relationships as accurately as we can. However, we cannot definitively confirm whether the evolutionary trees we build are correct — so scientists still grapple with how they should piece them together.
Biologists use two kinds of data to determine the relatedness of organisms: anatomy and appearance, which together are termed morphology, and DNA. In many cases, however, an evolutionary tree — also called a phylogeny — will branch differently if it is based on morphological similarities than if it is based on DNA. When this happens, which tree should we pick? There is no standard tree, and surprisingly, no study has evaluated two conflicting trees by using a third, independent means of measurement.
A team of researchers from the University of Bath, in Britain, sought to change that. The scientists evaluated morphological- and molecular-based phylogenies that show different relationships within the same group of organisms. The researchers used biogeography — the geographic distribution of species — as a test of the trees. Their results, published in the journal Communications Biology, show that molecular phylogenies matched with the geographic data significantly better than trees based on morphology. Their results demonstrate the utility of using geography to validate the determinations of evolutionary relationships. They further suggest that we may need to rebuild some evolutionary trees that use morphology rather than DNA to relate organisms to one another.
Look-alikes can be very different
Before the advent of molecular sequencing, scientists only considered anatomy when determining how animals might be related. This is far from ideal, because we know that looking alike does not always mean there is an evolutionary relationship between two species. When distantly related organisms face the same stressor, evolution tends to lead them toward similar solutions. Convergent evolution describes how distantly related organisms independently evolve the same traits. Consider our complex camera-type eye, which also developed independently in cephalopods such as the octopus. The wings of some dinosaurs, bats, and birds also evolved independently to serve a similar function. The same can be said for seemingly bizarre traits in plants, like carnivory or parasitism.
Convergent evolution occurs when animals must adapt to similar environmental stressors, or when they adopt similar ecological niches. For example, the flippers and gills of many marine animals evolved independently of one another to solve the problems of life underwater. Fundamental evolutionary theories such as those of Charles Darwin and Alfred Wallace rely on geographic patterns to back up evolutionary trees. But convergent evolution poses a problem for morphological trees: If distantly related organisms can evolve similar anatomical traits, how can we use morphology to assess whether organisms are related?
To determine the relationship between morphology and evolutionary history, the authors evaluated 48 pairs of evolutionary trees spanning a wide variety of organisms (such as terrestrial and freshwater vertebrates, insects, plants, and marine life). One of each pair was built on morphological data; the other on molecular data. When the authors mapped geographic distributions of the animals onto the trees, it became clear that the trees built on molecular data more closely matched distributions. In other words, family trees based on DNA more reliably predicted geographic distributions than morphology. Because we know that geographic distributions are related to evolutionary relationships, the authors concluded that family trees based on DNA more reliably depict evolutionary history.
Morphology’s enduring importance
Molecular data offers several advantages over morphology. First, DNA sequencing is easy, so a lot of data can be collected. Second, it relies less on taxonomic expertise. In some cases, only a few experts can tell two species apart based on subtle morphological differences. Consequently, it is much easier to compile large molecular data sets than it is to assemble equivalent volumes of morphological data.
Another problem is that morphology is too subjective. Different scientists might code the same feature differently. Molecular systematics, however, comes with rules that mitigate subjectivity and are mainly based on mathematical models.
Still, molecular sequencing has its problems. It is not a perfect science. Plus, in many cases, we cannot get DNA. The study’s authors stress that morphological data in phylogenetic analysis is still essential, especially since 98 percent of known species are extinct. For many of these species, we can only infer relatedness from fossils that have no DNA.
Our knowledge of how species are related will never be definitive — new methods may emerge that cause us to fundamentally rethink relationships between species. For now, when two conflicting phylogenies exist, the authors suggest that biologists prioritize molecular datasets. Importantly, we now know that we can use biogeographic tests to check evolutionary trees created using morphological, molecular, and combined data sets. As we begin to do so, we may redraw hundreds of classic evolutionary trees.