The next omics? Tracking a lifetime of exposures to better understand disease

In the summer of 2016, 100 teenage girls living in California’s Salinas Valley slipped silicone bracelets onto their wrists and committed to keep them there for a week. Around the same time, 92 preschoolers in Oregon accessorized the same way. The bands found their way onto the wrists of farmworkers in Peru and Houston residents working to rebuild after Hurricane Harvey. Aside from color, the wristbands looked much the same as the yellow Livestrong bracelets popularized by cyclist Lance Armstrong in the early 2000s.
But this was not some fashion trend shared by teenage girls, farmworkers and preschoolers. These bands were research tools, whose porous silicone made them ideal for soaking up chemicals in their surroundings. All were worn as part of a growing effort to understand just what is present in different environments and how those “exposures” — be they pesticides, smoke, floodwater contaminants or just the day-to-day contents of a preschool classroom — can affect health.
In homes, on buildings, from satellites and even in apps on the phone in your pocket, tools to monitor the environment are on the rise. At the intersection of public health and toxicology, these tools are fueling a new movement in exposure science. It’s called the exposome and it represents the sum of all environmental exposures over a lifetime. As explored in a trio of papers in the Annual Review of Public Health, scientists think that if we knew every substance that a person was exposed to at every moment ever, it could give rise to dramatic improvements in the understanding of the causes and risk factors for disease.
That goal, its architects will readily admit, is absurdly ambitious — even impossible. But even imperfectly realized, the approach may be what’s needed to finally understand why one person develops a disease and another doesn’t, which environmental exposures are the most worrying and if there are windows of vulnerability — times of life when exposures may be especially harmful.
“We think about all health and illness as a combination of genes and the environment, and now it really is time to fill out the environment side of that equation,” says Julia Brody, a toxicologist affiliated with Brown University and the leader of the Silent Spring Institute, which studies environmental links to breast cancer. “We’ve made an enormous investment in understanding the genome and almost nothing comparatively to focus on the exposome.”
Impossible or not, researchers are forging ahead, breaking off bits and pieces of a lifetime’s exposure to put under the microscope. Still, discussions continue about how to make real progress and how to collect and examine information in a meaningful way.
Some scientists are advocating for more air monitors in cities and homes. Others are developing wearable monitors that soak up pieces of the environment as people move through their day. Some are trying to match tracking data from cell phones to satellite indicators of air quality, helping to assess individual exposures based on a person’s locale and movements. Still other researchers are looking inside the body, hoping to identify chemical footprints that distinct exposures may have left behind.
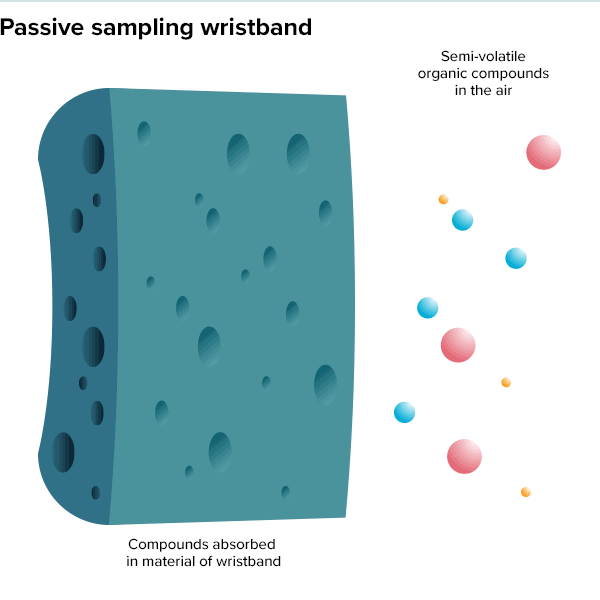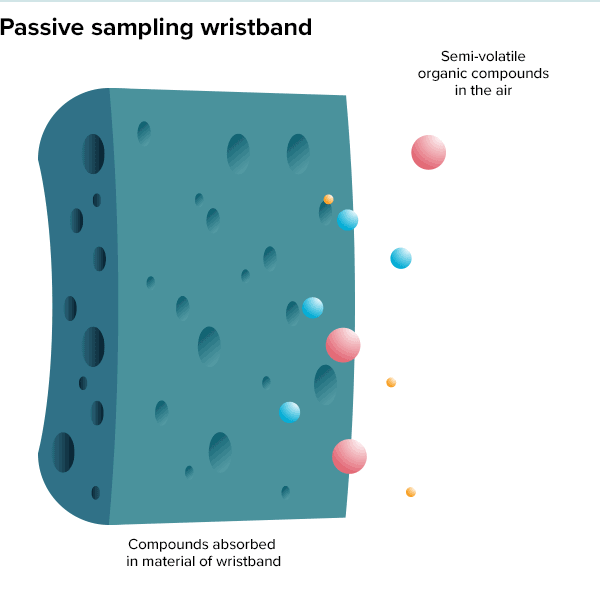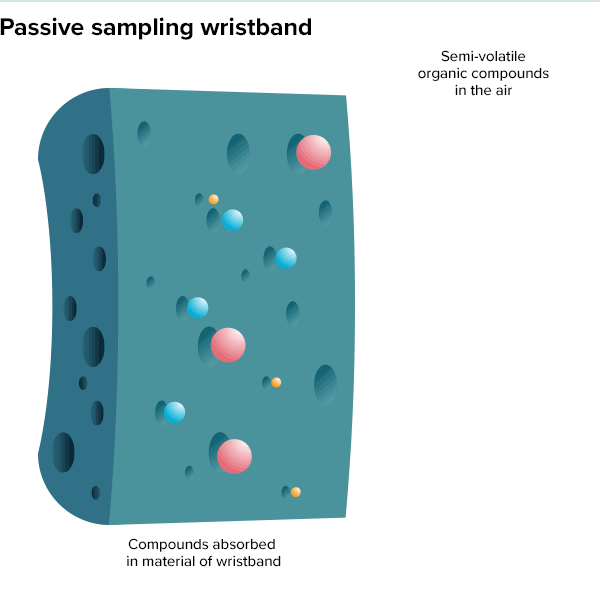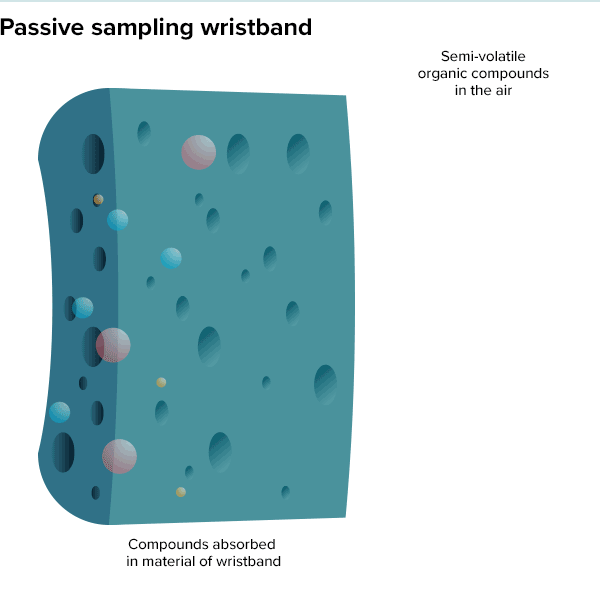
Though varied, the approaches share the goal of adding breadth to traditional exposure science studies, which have historically focused on linking a single disease to one or more suspicious exposures, often as cases build up over time. A history of poisoned lead workers dating back to the time of the Romans led modern researchers to demonstrate that even low levels of lead from automobile exhaust posed a serious public health risk. And a startling spike in lung cancer cases alongside increased smoking popularity drove researchers to perform focused studies on the dangers of cigarettes in the mid-twentieth century.
But many potentially harmful exposures could be far less obvious. The air you breathe, the food you eat, the products you use, the medicines you take, the surfaces you touch — all may contain multitudes of invisible chemicals (many naturally occurring, others not) and microbes you never know you’ve contacted. Thus, exposure science remains far from what it would ideally be: a guide to avoiding disease risk. Exposome scientists want to change that by capturing as broad a picture as possible. They want to ask not only if certain chemicals or microbes can harm health, but also if certain substances are dangerous in particular combinations, during particular times — such as during pregnancy — or to particular groups of people.
An “ome” of one’s own
It was a concern for unexplained diseases that led cancer researcher Christopher Wild to first come up with the term “exposome” in 2005. Wild had closely followed the race to sequence the human genome, which had successfully concluded two years prior, and worried that, in its eagerness to advance genetics, the world had forgotten the importance of environmental exposures in health. It’s a sentiment that has only grown stronger in recent years as genetics has failed to yield clear links to many cancers and other diseases. A recent study looked at the prevalence of 28 chronic conditions in twins and found that genetics explained less than 20 percent of the risk in most of the illnesses examined. Even in asthma — which ranked highest in terms of genetic contribution — genetics explained less than 50 percent of the risk. For leukemia — on the other end of the rankings — genetics explained only 3 percent.
“I was excited about the genetics,” says Wild, who formerly directed the World Health Organization’s International Agency for Research on Cancer. “But I felt it was an imbalance in the tools and the investment that we had available, and that was going to lead to problems. If we can’t measure this other component, we’re not going to get to the bottom of the problem.”
To remind the world that the genome was only half the equation, Wild coined an “ome” of his own. He hoped that if the two factors had mirroring names it would push other scientists to think of them as having equal importance. It took some time, but slowly his colleagues have begun to rally around the idea. Efforts to develop sophisticated devices that sample the world around us are on the rise. And new research groups are forming around the world to bring together the technology and expertise needed to process the vast amounts of data that will come from emerging exposomics projects.
One example is the Children’s Health Exposure Analysis Resource, a network of US laboratories and other resources established by the National Institute of Environmental Health Sciences to support researchers who want to add a stronger environmental component to their studies. CHEAR will allow scientists to test for more chemicals that children may be exposed to — hundreds to thousands at once — and will develop standardized testing to allow for data comparison across studies. The importance of such a resource is to help researchers break away from looking at a small number of potential threats at a time and start moving toward a way to take into account the entire realm of exposures, says Susan Teitelbaum, a member of CHEAR’s executive committee and a public health researcher at the Icahn School of Medicine at Mount Sinai in New York City.
Efforts so far have focused largely on establishing the technology, methods and collaborations needed to move forward, but early studies have already yielded some small insights. A preliminary study that used wristbands to examine chemical exposure in preschoolers found that, as they go about their daily lives, children are exposed to many flame retardants — even some that have been phased out due to concerns about toxicity.
Pilot studies to test new devices have led to a few, more personal realizations as well. Katherine Sward, a biomedical informatics researcher at the University of Utah, reported that a family trying out an in-home air monitor realized that vacuuming right before a visit from their child’s asthmatic friend might actually lead to more, not less, risk after the cleaning launched dust particles into the air. In another pilot, a researcher using a portable monitor to track exposure to airborne particles concluded that he was allergic to eucalyptus, not pine, as he previously believed, when he noted that his worst symptoms seemed to correlate with high levels of eucalyptus pollen detected by his device.
Looking inward
Researchers are still a long way from tracking even a fraction of the potential threats that surround us. Some think it might be more practical to tackle the problem by focusing on what’s inside the body, hunting for traces of past exposures. Signatures left over in blood, urine, teeth and even toenails can hint at previous exposures. Blood in particular holds clues that can let researchers work backwards to match biological changes to triggering exposures, says Dean Jones, a biochemist at Emory University in Atlanta and coauthor of a 2019 article about the promise of the exposome paradigm in the Annual Review of Pharmacology and Toxicology. The dream, he says, is to someday be able to create a readout of how the body is being affected by the environment and then determine the substances an individual should avoid, based on how they respond to each one.
“All people are pre-disease,” Jones says. “The vision is that we will have tools good enough to put data together into predictive programs and the predictive programs will say: You are likely to develop renal disease when you are in your 50s and if you do a, b, c, you’ll reduce that risk.”
In blood, Jones can look for metabolites, small molecules broken down or created by body processes. With tens of thousands of detectable components that hint at what the body is doing chemically, metabolites may be one possible alphabet scientists could use to read back what’s happened internally following various exposures.
At the UC Berkeley School of Public Health, Stephen Rappaport is exploring a different language found in blood. By examining human serum albumin — a protein that vacuums up damaging compounds circulating in blood — Rappaport can look for the residues those compounds have left behind. Both Jones and Rappaport think they can link these internal signs to external exposures. Eventually they aim to identify the ones that pose harm.
Blood can provide a snapshot of the goings-on inside the body, says Rappaport. “Most of the things we think about as being toxic or protective from disease are things that are actively transported by the blood.”
A data problem
Even if scientists can count on the body to point to harmful chemicals, other challenges remain. Once scientists have found something in the blood, they must try to trace it back to a known chemical in a database. But they often won’t find a match. The vast majority of the world’s chemicals (both natural and human-made) have never been characterized and new chemicals are created all the time.
Simply getting a handle on how many chemicals are out there is harder than it sounds. A recent study attempted this in 100 consumer products. Toothpaste contained 85 chemicals, while one plastic children’s toy contained about 300. Across all the products, the study detected 4,270 unique chemical signatures and tentatively identified 1,602 of those. But only 30 percent of those 1,602 chemicals could be matched to public lists of known ingredients in consumer products or compounds of toxicological interest.
“There’s got to be tens of millions — hundreds of millions, if not more,” says Jon Sobus, an environmental health scientist at the US Environmental Protection Agency and coauthor on the consumer products study. “Where does the number of chemicals end?”
With so many unknowns, scientists are left with a huge number-crunching issue, making the exposome, like so many fields today, a big data problem. That’s forcing scientists to devise new ways to analyze the information they collect. One approach gaining popularity borrows from a common, if sometimes controversial, method in genetics. Dubbed the genome-wide association study, or GWAS, the method looks to see which of thousands of genes vary in conjunction with a particular disease or symptom.
In 2010, Harvard bioinformatician Chirag Patel adapted the GWAS into an environment-wide association study to see how 266 environmental factors varied in step with the risk of developing type II diabetes. The study rediscovered certain factors already linked to diabetes risk, but also pointed researchers to other potential risks, including a dietary component of vitamin E and heptachlor, a pesticide previously linked to type II diabetes in workers who applied it, but not known to have an effect at smaller levels in the general population.
After this, other researchers adopted the EWAS method to look for substances that might confer risk and should be studied further. In a 2011 study examining metabolites in human plasma, researchers found three small molecules that correlated with increased risk of cardiovascular disease later on. One of these was trimethylamine N-oxide, or TMAO. TMAO is a byproduct of choline, which is found in certain foods, including meat and eggs. Researchers realized that certain gut microbes converted choline to an intermediate compound, which was then broken down to TMAO in the liver. In mice, high TMAO levels corresponded to thickened artery walls. The researchers proposed that manipulating a patient’s microbiome might be able to protect people from this type of dietary exposure.
The study was applauded by researchers like Rappaport, who sees it as a validation of the EWAS method. Still GWAS itself is far from perfect and the EWAS faces many additional challenges when compared to its genetic predecessor. A genome-wide study is contained in a way that an environment-wide search is not. Humans have a lot of genes — about 20,000 that code for proteins. But that number pales in comparison to the number of things people might encounter in the environment. And unlike the genome — which is packaged together in a place we know how to find — our “exposures” can be anywhere at any time in any quantity.
The sheer scope of the problem is daunting to researchers just hearing about the exposome. But for Sobus, it’s not unlike the early days of genomics, when many wondered if mapping the human genome was an unfeasible goal. As more tools and resources are created, more scientists are finding ways to incorporate the idea into their studies, he says. And each researcher who decides to tackle even a small chunk helps move the field forward.
“I think everyone kind of goes through that growth period of a little bit of disbelief,” he says. “Then you start warming up to it and you want to get on the band wagon.”
This article originally appeared in Knowable Magazine, a nonprofit publication dedicated to making scientific knowledge accessible to all. Sign up for Knowable Magazine’s newsletter.