Scientists can now map molecular structures in minutes
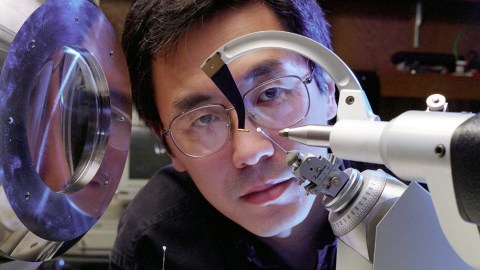
Dr. Ming Luo, associate director of the Center for Macromolecular Crystallography, X-rays flu virus crystals grown in space to map its structure for drug developers. Photo credit: Nasa/Getty Images
- X-ray crystallography allows scientists to accurately map molecular structures, but the process is slow thanks to the need to grow sufficiently sized crystals.
- Two independent research teams have discovered a way to use electron diffraction to accurately map molecular structures with incredibly small samples.
- The results of their research have been published in both Angwandte Chemie and ChemRxiv.
People tend to think of x-rays, if they think of them at all, as a quick-and-easy way to snapshot hidden worlds. You sit back, the dentist throws a lead-heavy vest on you, and the x-rays go to work. Voila! You have an atomic rendering of your molar. Chemists use x-rays to map out molecular structures, but in the microscopic world, x-rays prove less than the insta-images we think they are. They aren’t even Polaroid. They are more ye olde portait, taking weeks or even months to completely map out a single crystal structure.
But that’s about to change. Two independent research teams have found a way to use electrons to map crystal structures in mere minutes, significantly reducing the wait time. Last month, the teams published their work in Angewandte Chemie and ChemRxiv*. “I haven’t been this excited about a finding in chemistry in a long time,” Donna Huryn, an organic and medicinal chemist at the University of Pittsburgh, told Science News. “It’s going to change the way everybody works.”

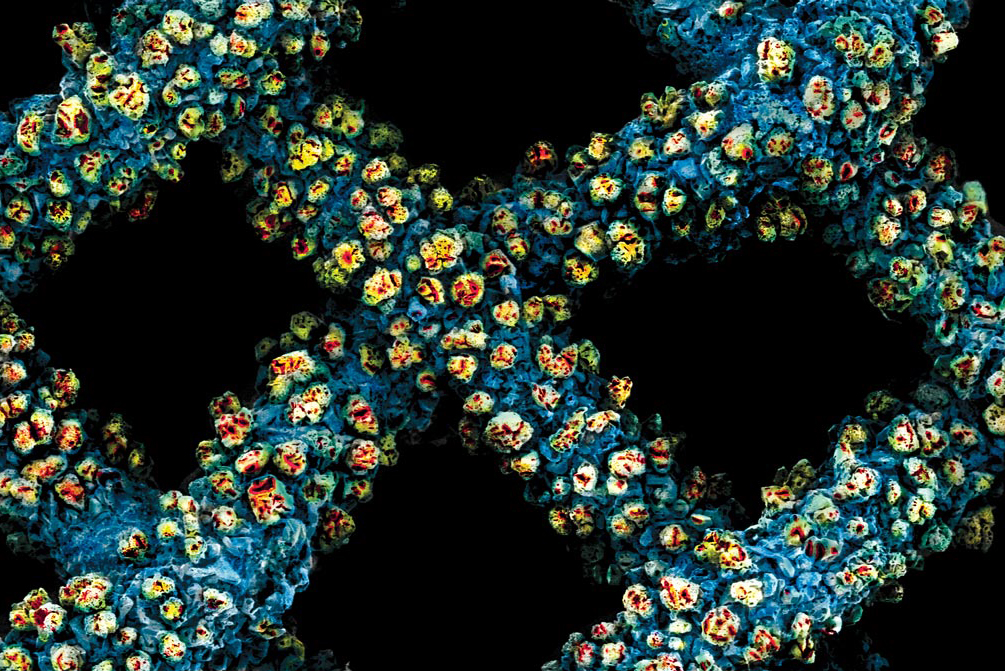
X-ray crystallography of a protein crystal. Image source: CSIRO
Molecular paint by numbers
According to the Angewandte Chemie paper, chemists publish about 50,000 crystal structures a year. The majority of these are created using x-ray crystallography (also known as x-ray diffraction). During this process x-rays are shot at a crystal structure and bend, or diffract, when striking it. By measuring how the x-rays diffract off the crystal, chemists can map out its structure to determine its makeup and various characteristics. This allows chemists to determine the compositions of the substances they work with.
However, as the authors of the ChemRxiv paper note, it is a technique with limitations.
“The process is considered an art, where the production of high quality [sic] crystals suitable for x-ray diffraction requires uncodified ‘tricks of the trade’ and a certain amount of luck!” they write. “Additionally, even once a substance has been successfully crystallized, there is no guarantee that the particular crystal form will be amenable to x-ray diffraction.” [Emphasis original]
Another issue, cited by both papers, is that it can take a long time to grow crystals. And as anyone who has ever created a sugar crystal structure for the science fair can tell you, it doesn’t always turn out. Some scientists can spend their careers learning to build quality crystals of a large enough size.
Growing crystals for x-ray crystallography is the main reason it can take so long to map out a single molecular structure.
Speeding things up with electrons
To get around this size issue, the two teams used electrons instead. Electrons diffract more strongly than x-rays, creating higher-resolution images. More importantly, the size of the crystal doesn’t have to be near as large — just that of a “grain of icing sugar” according to the Angewandte paper.
After successes in identifying and mapping crystal structures, both research teams pushed the boundaries of electron diffraction by testing it on heterogenous samples (that is, compound mixtures).
For the Angewandte paper, the researchers looked at cold medicine containing both active and inactive ingredients and were able to suss out the structure of the active ingredient (acetaminophen). In the ChemRxiv paper, the researchers crushed four compounds together and analyzed the mixture. They were able to separately map out the structure of each compound.
As noted by Carmen Drahl at Science News, using electrons to map structures isn’t new. The 2017 Nobel Prize in chemistry went to scientist who mapped proteins with the technique. However, the fact that chemists may now be able to use this technique to map molecular structures from samples so small is a real game changer.

DNA as seen through x-ray crystallography. Image source: Wikimedia Commons
A new century of discoveries
During World War II, chemist Dorothy Hodgkin and her colleagues used x-ray crystallography to determine the structure of penicillin. With this structure, chemists were able to mass-produce a synthetic version of the drug for wartime efforts. The same team also worked on mapping vitamin B12 and insulin.
Electron diffraction may be used to similar medical benefits but today in a fraction of the time. It could also be used to analyze drug purity and understand the intricacies of virus particles, thanks to its ability to achieve high-resolution images using minute samples.
* Please note: Angewandte Chemie is a peer-reviewed scientific journal, while ChemRxiv houses manuscript preprints before they are submitted for peer review. As such, some of the material quoted from the ChemRxiv paper may change by the time it reaches peer-reviewed publication.