‘Micro snails’ we scraped from sidewalk cracks help unlock details of ancient Earth’s biological evolution

Every step you take, you’re likely walking on a world of unseen and undescribed microbial diversity. And you don’t need to head out into nature to find these usually unnoticed microscopic organisms.
Asbiologists, we know this firsthand. A meetup for coffee several years ago ended with our using makeshift sampling tools – actually a coffee stirrer and a coffee cup lid – to collect some of the black gunk from between the sidewalk’s concrete slabs. In this mundane space on the Mississippi State University campus, we discovered microbes that have lived on Earth for millions of years.
Finding these charismatic organisms in the environment, while exciting, is just the first step. Our mutual interest is to better understand how organisms are related to one another. We’re using DNA to reveal their relationships in the very distant past.
By sampling organisms that are alive today, we can ask deeper questions about the evolution that happened millions of years ago in now extinct ancestors.
Piecing together the tree of life
Our simple act of collection after our 2015 coffee date started a fruitful collaboration betweenour labs in the field of molecular protistology. Our focus is on the microscopic single-celled organisms called protists, particularly ones that move around using tiny tentacles called pseudopodia.
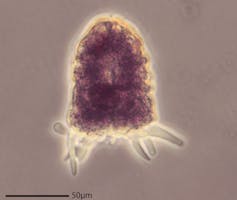
Amphizonella – identified in the authors’ sidewalk sample – has a soft protective layer.
Matthew W. Brown, CC BY-ND
One elusive critter we identified in our sidewalk sample is an amoeba named Amphizonella; we joke that it makes its own “leather jacket” in the form of a soft, protective outer layer.
Despite what other scientists had previously thought, we had a hunch that this organism wasn’t closely related to other amoebae that have tougher outer coverings. This other much larger group, called testate amoebae, have shells – imagine microscopic snails – instead of leather jackets.
Because testate amoebae make a hard shell, they have the potential to fossilize. In fact, their vivid fossil record represents some of the oldest unequivocal fossils of eukaryotes – the category of life whose members hold their DNA within their cells’ nuclei. Why is this important? Human beings are also eukaryotes, as are plants, fungi, other animals, kelps and protists. Because these amoebae are some of the oldest eukaryotic fossils, they can in turn tell researchers like us something about our own species’ origins.
Since the advent of DNA sequencing in the early 2000s, biologists have used a small piece of the genome, even a single gene, to examine the relationships between organisms, though with limited success. Through similarity of DNA sequences between living organisms, one can infer relationships using complex computational approaches that model evolution change over time from empirically derived data. Simply put, scientists try to piece together who’s related to whom in order to reconstruct the evolutionary tree of life, or what we call a phylogenetic tree.
The first step of single-cell transcriptomics is isolating a single organism. Here, a micropipette picks up one Amphizonella cell. Credit: Matthew W. Brown.
In most cases the testate amoebae are quite difficult to cultivate in the laboratory, making it very hard to obtain enough material to sequence their DNA with the usual methods.
To overcome these challenges, we’re using a cutting-edge technique that allows us to take an organism directly from the environment and sequence its entire transcriptome – that’s the blueprint of all the proteins that it makes. This way, we’re able to bypass sequencing the whole genome (with its extraneous information) and sequence only the protein-coding regions. We end up with high-quality data of billions of base pairs of DNA that we can directly compare with similar data from other organisms.
This method provides better resolution by sampling hundreds of genes, rather than a single one. Then we use the data to build a phylogenetic tree of life that organizes our amoebae by how closely related they are to each other based on the similarity of their DNA. With these data, we can go further and compare our testate amoebae to other eukaryotes and identify what makes them unique and similar at a genomic level.
Connecting today’s life to ancient ancestors
Because life evolved over billions of years from a last universal common ancestor, all organisms, both living and extinct, must be related to each other in a single family tree.
But fossils don’t preserve DNA information. While it’s true that some ancient DNA sequencing is possible, in general it’s only been done with frozen samples like the woolly mammoth or ancient human beings like mummified remains. These ancient DNA samples have not really fossilized, and in comparison to fossils, they’re significantly more recent – for instance, the oldest human-related DNA to have been sequenced was from a Denisovan person’s tooth, which is about 110,000 years old.
In contrast, the fossil of Archaeopteryx, one of the most ancient relatives of birds, is about 150 million years old. That means that, today, we are about 100,000 times more distant to Archaeopteryx than we are to the Denisovan remains. That’s an immense amount of time.
A scanning electron micrograph of a fossilized Ciclocyrillium torquata, sampled from the Urucum formation in central Brazil.
Luana Morais, CC BY-ND
The fossils that seem to relate to today’s testate amoebae are about 750 million years old, from a time period called the Neoproterozoic. Scientists know very little about what was happening on Earth in that very distant past. Researchers have identified these tiny fossils in rocks collected in the Grand Canyon and central Brazil.
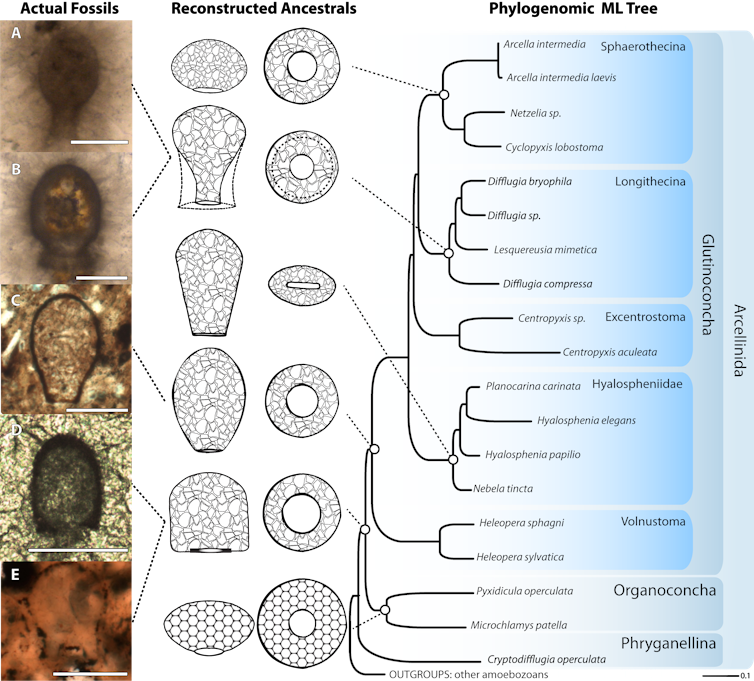
In order to compare the tree we created based on DNA from living species with the fossilized shells of the Neoproterozoic, we had to somehow extrapolate our data. Using the rates of evolution calculated in our tree, we were able to apply these rates using how the shells look today, to estimate what they might have looked in the past. This way, we can create a hypothetical ancestor that we can then compare to the actual fossils.
A family tree of testate amoebae linking the fossil record (left) to present day testate amoebae (right).
Lahr et al. 2019, Current Biology https://doi.org/10.1016/j.cub.2019.01.078., CC BY-ND
Our results were impressive. We calculated seven hypothetical ancestors based on a few million possibilities. When we compared them to the fossil record, previously described in the literature, we found five fossil species that were incredibly similar to our predictions. This allowed us to confidently determine that those Neoproterozoic fossils are indeed very ancient testate amoebae, and that this group has been around since before 750 million years ago. And even by then, they had already considerably diversified.
Showing that these creatures were around and diverse at such deep time scales is important because they’re complex organisms, with complex ecologies and behaviors. They provide an inside look into what life might have been like in those ancient eras. The amoebae can be predators, but they can also be grazers, or even harbor symbiotic algae that produce their food, making them primary producers.
The fact that many diverse types of testate amoebae were around by this stage implies that complex food webs had already developed, which in turn has implications for what the environment might have been like. Now, geochemists will compare their notes to our biological insights, and our understanding of ancient earth will continue to improve.![]()
Matthew Brown, Assistant Professor of Biological Sciences, Mississippi State University and Daniel Lahr, Assistant Professor of Zoology, Universidade de São Paulo
This article is republished from The Conversation under a Creative Commons license. Read the original article.